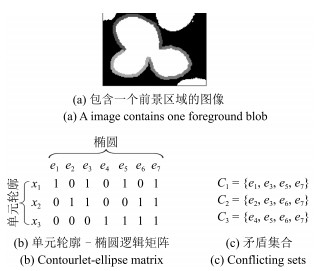
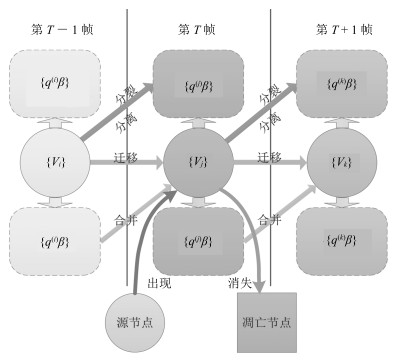
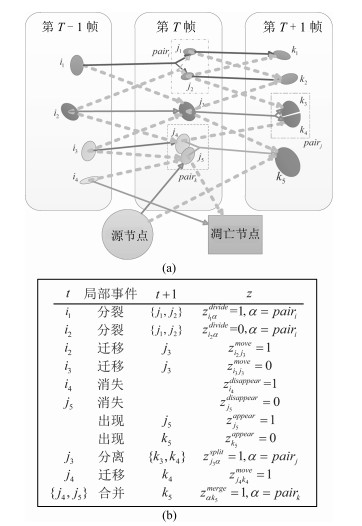
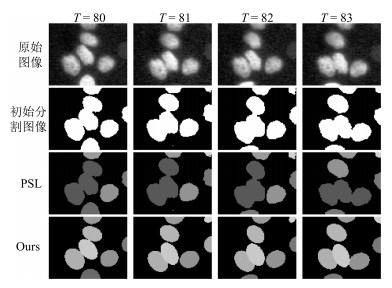
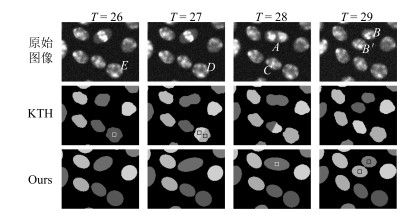
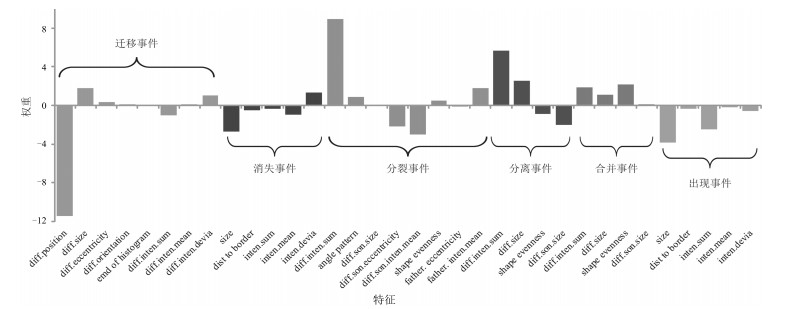
-
摘要: 提出一种新的多细胞联合检测与跟踪方法,通过椭圆拟合构建细胞观测假说的完备集合,定义了多种局部事件来描述细胞的行为以及检测阶段可能出现的错误.通过引入相应的标签变量,将细胞跟踪建模为结构化预测问题,通过求解一个带约束的整数规划问题得到细胞轨迹的全局最优解.针对结构化预测模型中的参数学习问题,本文采用Block-coordinate Frank-Wolfe优化算法根据给定的训练样本求解模型的最优参数,同时给出了该算法的非线性核化版本.本文在多个公开数据集上对提出的算法进行了验证,结果表明,本文的实验表现相比于传统算法有着显著的提升.
-
关键词:
- 细胞跟踪 /
- 结构化预测 /
- 结构化学习 /
- Block-coordinate Frank-Wolfe算法
Abstract: In this work we propose a new joint detection and tracking method for cell tracking. First we develop a new procedure for generating an over complete set of detection hypothesis via ellipse fitting methods. Then we define several local events and corresponding labeling variables to account for the biological behavior of cells and the imperfection in segmentation, and formulate the task of cell tracing as an integer programming problem with constraints. In addition, instead of learning local classifiers, we exploit a recently proposed block-coordinate Frank-Wolfe algorithm to automatically learn optimal parameters of our model. We also present the kernelized version of the learning algorithm which can boost the tracking performance even further. We conduct extensive experiments on public datasets, showing that our method consistently outperforms traditional countetparts.1) 本文责任编委 贺威 -
表 1 数据集的相关统计信息
Table 1 Statistics of dataset used in our study
数据集名称 训练图像序列长度 测试图像序列长度 图像大小 初始分割F-值 (%) Fluo-N2DL-HeLa-01 92 92 1100×700 94.2 Fluo-N2DL-HeLa-02 92 92 1100×700 92.7 Fluo-N2DH-SIM+-01 65 130 660×718 96.4 Fluo-N2DH-SIM+-02 150 138 664×790 95.7 表 2 本文算法与当前几种最好的算法之间的比较 (%)
Table 2 Comparison of our algorithm against state-of-the-art methods (%)
迁移事件 分裂事件 全部事件 精度 召回率 F-值 精度 召回率 F-值 精度 召回率 F-值 HeLa-1 KTH 98.1 96.4 97.2 75.3 74.8 75.1 97.4 95.3 96.3 NFP 98.9 99.1 99.0 85.1 76.2 80.4 98.1 98.1 98.1 EPFL 97 99 98 92 79 85 N/A N/A N/A PSL 97.7 91.8 94.7 82.7 73.4 77.8 94.8 91.6 93.1 Ours-P 98.2 98.7 98.5 92.1 74.1 82.1 97.2 98.2 97.7 Ours 99.1 99.3 99.2 88.5 86.0 87.2 97.9 98.2 98.1 HeLa-2 KTH 95.1 98.3 96.7 76.3 77.5 76.9 94.7 97.5 96.1 NFP 98.0 98.3 98.2 84.5 84.5 84.5 96.1 96.4 96.3 EPFL 96 99 97 86 83 84 N/A N/A N/A PSL 96.8 90.7 93.7 73.6 73.6 73.6 94.3 90.1 92.2 Ours-P 97.3 97.9 97.6 82.8 86.0 84.4 96.1 97.3 96.7 Ours 97.9 98.6 98.3 92.8 89.9 91.3 97.2 98.0 97.6 SIM+-01 KTH 97.7 98.9 98.3 73.8 51.7 60.8 96.8 97.9 97.3 NFP 99.1 96.8 98.0 76.1 85.0 80.3 97.3 96.6 97.0 EPFL N/A N/A N/A N/A N/A N/A N/A N/A N/A PSL 98.9 94.3 96.7 33.3 73.3 45.8 91.7 94.1 92.9 Ours-P 99.7 96.5 98.1 60.9 93.3 73.7 93.6 96.5 95.0 Ours 99.8 99.9 99.9 94.6 89.8 92.2 99.7 99.8 99.7 SIM+-02 KTH 94.4 92.6 93.5 80.2 72.7 76.3 93.2 92.1 92.6 NFP 97.1 90.0 93.4 79.6 81.8 80.7 94.4 91.1 92.7 EPFL N/A N/A N/A N/A N/A N/A N/A N/A N/A PSL 96.4 94.1 95.2 73.8 84.1 78.6 94.7 93.6 94.1 Ours-P 96.4 94.0 95.2 82.0 86.4 84.1 95.0 93.6 94.3 Ours 97.6 93.9 95.7 86.4 90.1 88.2 96.6 93.6 95.1 表 3 HeLa数据集上训练样本长度对于训练和预测的影响 (%)
Table 3 Effects of training sample length on training and prediction HeLa (%)
数据集 样本数量 单个样本长度 训练时间 (min) 训练误差 迁移事件F-M 分裂事件F-M 全部事件F-M HeLa-1 1 80 101.5 3.5 99.3 86.8 98.9 2 40 41.0 2.1 99.2 82.5 98.1 4 20 25.3 2.0 99.3 87.5 98.9 8 10 14.6 1.7 99.1 87.7 98.7 10 8 15.4 10.3 98.6 72.5 98.0 16 5 12.3 10.6 98.9 77.8 98.3 20 4 4.4 10.7 98.5 69.7 97.7 HeLa-2 1 80 96.3 4.3 98.1 86.2 97.1 2 30 28.8 4.8 97.8 82.7 96.7 4 15 8.9 5.2 97.9 90.4 97.1 6 10 4.8 4.8 98.3 91.3 97.6 10 6 4.0 6.0 98.3 89.4 97.5 15 4 1.8 6.4 98.1 89.7 97.0 20 3 1.1 7.3 95.9 55.3 93.6 表 4 SIM+-01上的核化学习效果 (%)
Table 4 Effects of kernelization SIM+-01 (%)
核函数类型 参数 训练时间 (min) 迁移事件 分裂事件 精度 召回率 F-值 精度 召回率 F-值 RBF 100 5.8 99.8 100 99.9 94.6 88.1 91.2 10 4.5 99.8 100 99.9 92.9 88.1 90.4 1 4.5 99.8 100 99.9 98.2 91.5 94.7 0.1 4.8 99.8 100 99.9 96.4 91.5 93.9 0.01 4.3 99.8 100 99.9 96.4 91.5 93.9 0.001 5.0 99.9 100 99.9 98.3 94.9 96.6 Linear 2.0 99.8 99.9 99.9 94.6 89.8 92.2 表 5 在HeLa-1数据集上加入可选约束的效果
Table 5 Effects of optional constraints on HeLa-1 dataset
细胞局部事件
(真实发生数目)迁移
(12709)分裂
(143)分离
(1)TP FP TP FP TP FP 不加可选约束 12575 115 119 20 1 12 加入可选约束 12590 119 125 17 1 0 -
[1] Krzic U, Gunther S, Saunders T E, Streichan S J, Hufnagel L. Multiview light-sheet microscope for rapid in toto imaging. Nature Methods, 2012, 9(7): 730-733 doi: 10.1038/nmeth.2064 [2] Tomer R, Khairy K, Amat F, Keller P J. Quantitative high-speed imaging of entire developing embryos with simultaneous multiview light-sheet microscopy. Nature Methods, 2012, 9(7): 755-763 doi: 10.1038/nmeth.2062 [3] Meijering E, Dzyubachyk O, Smal I, van Cappellen W A. Tracking in cell and developmental biology. Seminars in Cell and Developmental Biology, 2009, 20(8): 894-902 doi: 10.1016/j.semcdb.2009.07.004 [4] Kanade T, Yin Z Z, Bise R, Huh S, Eom S, Sandbothe M F, Chen M. Cell image analysis: algorithms, system and applications. In: Proceedings of the 2011 IEEE Workshop on Applications of Computer Vision (WACV). Kona, HI: IEEE, 2011. 374-381 [5] Meijering E, Dzyubachyk O, Smal I. Methods for cell and particle tracking. Methods in Enzymology, 2012, 504: 183-200 doi: 10.1016/B978-0-12-391857-4.00009-4 [6] González G, Fusco L, Benmansour F, Fua P, Pertz O, Smith K. Automated quantification of morphodynamics for high-throughput live cell time-lapse datasets. In: Proceedings of the 10th IEEE International Symposium on Biomedical Imaging. San Francisco, CA: IEEE, 2013. 664-667 [7] Maška M, Ulman V, Svoboda D, Matula P, Matula P, Ederra C, Urbiola A, Espaňa T, Venkatesan S, Balak D M W, Karas P, Bolcková T, Štreitová M, Carthel C, Coraluppi S, Harder N, Rohr K, Magnusson K E G, Jaldén J, Blau H M, Dzyubachyk O, Křížek P, Hagen G M, Pastor-Escuredo D, Jimenez-Carretero D, Ledesma-Carbayo M J, Muñoz-Barrutia A, Meijering E, Kozubek M, Ortiz-de-Solorzano C. A benchmark for comparison of cell tracking algorithms. Bioinformatics, 2014, 30(11): 1609-1617 doi: 10.1093/bioinformatics/btu080 [8] 李振兴, 刘进忙, 李松, 白东颖, 倪鹏.基于箱式粒子滤波的群目标跟踪算法.自动化学报, 2015, 41(4): 785-798 http://www.aas.net.cn/CN/abstract/abstract18653.shtmlLi Zhen-Xing, Liu Jin-Mang, Li Song, Bai Dong-Ying, Ni Peng. Group targets tracking algorithm based on box particle filter. Acta Automatica Sinica, 2015, 41(4): 785-798 http://www.aas.net.cn/CN/abstract/abstract18653.shtml [9] 杨峰, 王永齐, 梁彦, 潘泉.基于概率假设密度滤波方法的多目标跟踪技术综述.自动化学报, 2013, 39(11): 1944-1956 doi: 10.3724/SP.J.1004.2013.01944Yang Feng, Wang Yong-Qi, Liang Yan, Pan Quan. A survey of PHD filter based multi-target tracking. Acta Automatica Sinica, 2013, 39(11): 1944-1956 doi: 10.3724/SP.J.1004.2013.01944 [10] 何涛, 张家树, 华见, 龚小彪.基于单固定摄像头的多目标跟踪框架.计算机应用研究, 2015, 32(6): 1892-1896 http://www.cnki.com.cn/Article/CJFDTOTAL-JSYJ201506068.htmHe Tao, Zhang Jia-Shu, Hua Jian, Gong Xiao-Biao. Multi-target tracking framework for fixed camera. Application Research of Computers, 2015, 32(6): 1892-1896 http://www.cnki.com.cn/Article/CJFDTOTAL-JSYJ201506068.htm [11] Alexandre D, Vasily S, Shahragim T, Dufour A, Shinin V, Tajbakhsh S, Guillen-Aghion N, Olivo-Marin J C, Zimmer C. Segmenting and tracking fluorescent cells in dynamic 3-D microscopy with coupled active surfaces. IEEE Transactions on Image Processing, 2005, 14(9): 1396-1410 doi: 10.1109/TIP.2005.852790 [12] Li K, Chen M, Kanade T, Miller E D, Weiss L E, Campbell P G. Cell population tracking and lineage construction with spatiotemporal context. Medical Image Analysis, 2008, 12(5): 546-566 doi: 10.1016/j.media.2008.06.001 [13] Dzyubachyk O, van Cappellen W A, Essers J, Niessen W J, Meijering E. Advanced level-set-based cell tracking in time-lapse fluorescence microscopy. IEEE Transactions on Medical Imaging, 2010, 29(3): 852-8 67 doi: 10.1109/TMI.2009.2038693 [14] Dufour A, Thibeaux R, Labruyere E, Guillen N, Olivo-Marin J C. 3-D active meshes: fast discrete deformable models for cell tracking in 3-D time-lapse microscopy. IEEE Transactions on Image Processing, 2011, 20(7): 1925-1937 https://www.researchgate.net/publication/224208247_3-D_Active_Meshes_Fast_Discrete_Deformable_Models_for_Cell_Tracking_in_3-D_Time-Lapse_Microscopy [15] Delgado-Gonzalo R, Chenouard N, Unser M. Fast parametric snakes for 3D microscopy. In: Proceedings of the 9th IEEE International Symposium on Biomedical Imaging (ISBI). Barcelona, Spain: IEEE, 2012. 852-855 [16] Maška M, Daněk O, Garasa S, Rouzaut A, Munoz-Barrutia A, Ortiz-de-Solorzano C. Segmentation and shape tracking of whole fluorescent cells based on the Chan-Vese model. IEEE Transactions on Medical Imaging, 2013, 32(6): 995-1006 doi: 10.1109/TMI.2013.2243463 [17] Amat F, Lemon W, Mossing D P, McDole K, Wan Y, Branson K, Myers E W, Keller P J. Fast, accurate reconstruction of cell lineages from large-scale fluorescence microscopy data. Nature Methods, 2014, 11(9): 951-958 https://www.researchgate.net/publication/264092799_Fast_accurate_reconstruction_of_cell_Lineages_from_Large-scale_fluorescence_microscopy_data [18] Chenouard N, Bloch I, Olivo-Marin J C. Multiple hypothesis tracking for cluttered biological image sequences. IEEE Transactions on Pattern Analysis and Machine Intelligence, 2013, 35(11): 2736-3750 doi: 10.1109/TPAMI.2013.97 [19] Li F H, Zhou X B, Ma J W, Wong S T C. Multiple nuclei tracking using integer programming for quantitative cancer cell cycle analysis. IEEE Transactions on Medical Imaging, 2010, 29(1): 96-105 http://citeseerx.ist.psu.edu/viewdoc/summary?doi=10.1.1.383.2134 [20] Kausler B X, Schiegg M, Andres B, Lindner M, Koethe U, Leitte H, Wittbrodt J, Hufnagel L, Hamprecht F A. A discrete chain graph model for 3d+t cell tracking with high misdetection robustness. In: Proceedings of the 12th European Conference on Computer Vision. Florence, Italy: Springer, 2012. 144-157 [21] Luo X H, Schiegg M, Hamprecht F A. Active structured learning for cell tracking: algorithm, framework, and usability. IEEE Transactions on Medical Imaging, 2014, 33(4): 849-860 [22] Magnusson K E G, Jaldén J. A batch algorithm using iterative application of the Viterbi algorithm to track cells and construct cell lineages. In: Proceedings of the 9th IEEE International Symposium on Biomedical Imaging (ISBI). Barcelona, Spain: IEEE, 2012. 382-385 [23] Padfield D, Rittscher J, Roysam B. Coupled minimum-cost flow cell tracking for high-throughput quantitative analysis. Medical Image Analysis, 2011, 15(4): 650-668 doi: 10.1016/j.media.2010.07.006 [24] Schiegg M, Hanslovsky P, Kausler B X, Hufnagel L, Hamprecht F A. Conservation tracking. In: Proceedings of the 2013 IEEE International Conference on Computer Vision. Sydney, Australia: IEEE, 2013. 2928-2935 [25] Schiegg M, Hanslovsky P, Haubold C, Koethe U, Hufnagel L, Hamprecht F A. Graphical model for joint segmentation and tracking of multiple dividing cells. Bioinformatics, 2015, 31(6): 948-956 doi: 10.1093/bioinformatics/btu764 [26] Magnusson K E G, Jaldén J, Gilbert P M, Blau H M. Global linking of cell tracks using the viterbi algorithm. IEEE Transactions on Medical Imaging, 2015, 34(4): 911-929 doi: 10.1109/TMI.2014.2370951 [27] Türetken E, Wang X C, Becker C J, Haubold C, Fua P. Network flow integer programming to track elliptical cells in time-lapse sequences. IEEE Transactions on Medical Imaging, 2017, PP(99): 1-1, DOI: 10.1109/TMI.2016.2640859 [28] Lacoste-Julien S, Jaggi M, Schmidt M, Pletscher P. Block-coordinate Frank-Wolfe optimization for structural SVMs. Eprint Arxiv, 2012, 53-61 [29] Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez J Y, White D J, Hartenstein V, Eliceiri K, Tomancak P, Cardona A. Fiji: an open-source platform for biological-image analysis. Nature Methods, 2012, 9(7): 676-682 doi: 10.1038/nmeth.2019 [30] Kovesi P. MATLAB and Octave Functions for Computer Vision and Image Processing [Online], available: http://www.peterkovesi.com/matlabfns/, November 3, 2015 [31] Prasad D K, Quek C, Leung M K H, Cho S Y. A parameter independent line fitting method. In: Proceedings of the 1st Asian Conference on Pattern Recognition. Beijing, China: IEEE, 2011. 441-445 [32] Prasad D K, Leung M K H, Quek C. ElliFit: an unconstrained, non-iterative, least squares based geometric ellipse fitting method. Pattern Recognition, 2013, 46(5): 1449-1465 doi: 10.1016/j.patcog.2012.11.007 [33] Nowozin S, Lampert C H. Structured learning and prediction in computer vision. Foundations and TrendsoledR in Computer Graphics and Vision, 2010, 6(3-4): 185-365 doi: 10.1561/0600000033 [34] de Solórzano C O, Kozubek M, Meijering E, Barrutia A M. ISBI Cell Tracking Challenge [Online], available: http://www.codesolorzano.com/celltrackingchallenge/CellTrackingChallenge/Welcome.html, November 15, 2015 [35] Lófberg J. YALMIP: a toolbox for modeling and optimization in MATLAB. In: Proceedings of the 2004 IEEE International Symposium on Computer Aided Control Systems Design. Taipei, China: IEEE, 2004. 284-289 [36] Bensch R, Ronneberger O. Cell segmentation and tracking in phase contrast images using graph cut with asymmetric boundary costs. In: Proceedings of the 12th IEEE International Symposium on Biomedical Imaging. New York, United States: IEEE, 2015. 1220-1223 [37] Achanta R, Shaji A, Smith K, Lucchi A, Fua P, Süsstrunk S. SLIC superpixels compared to state-of-the-art superpixel methods. IEEE Transactions on Pattern Analysis and Machine Intelligence, 2012, 34(11): 2274-2282 doi: 10.1109/TPAMI.2012.120 -




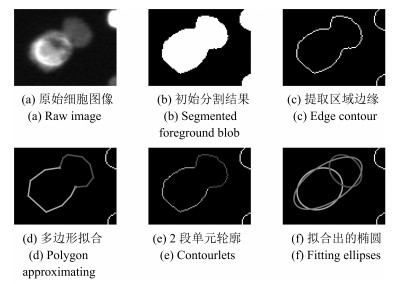
 下载:
下载: