-
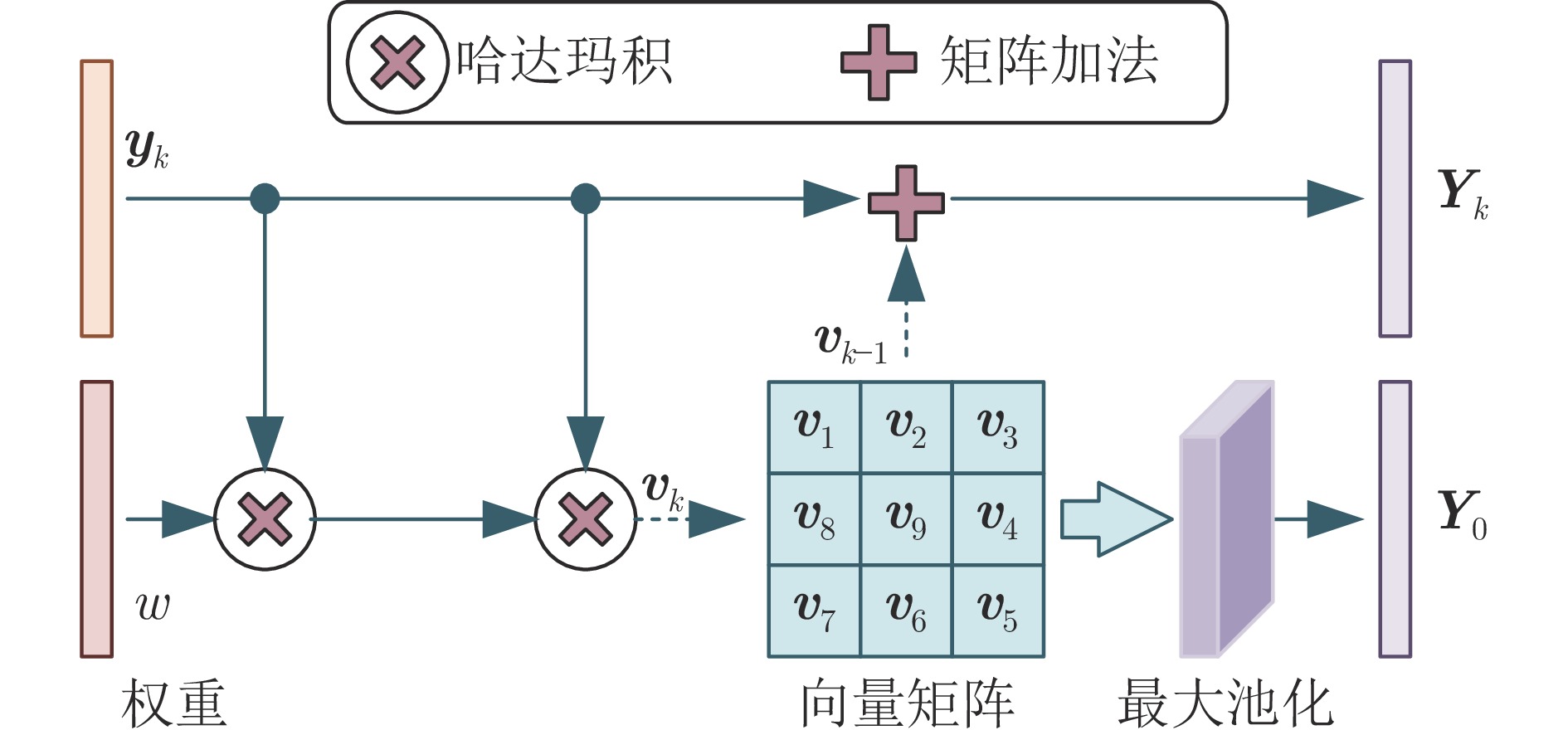
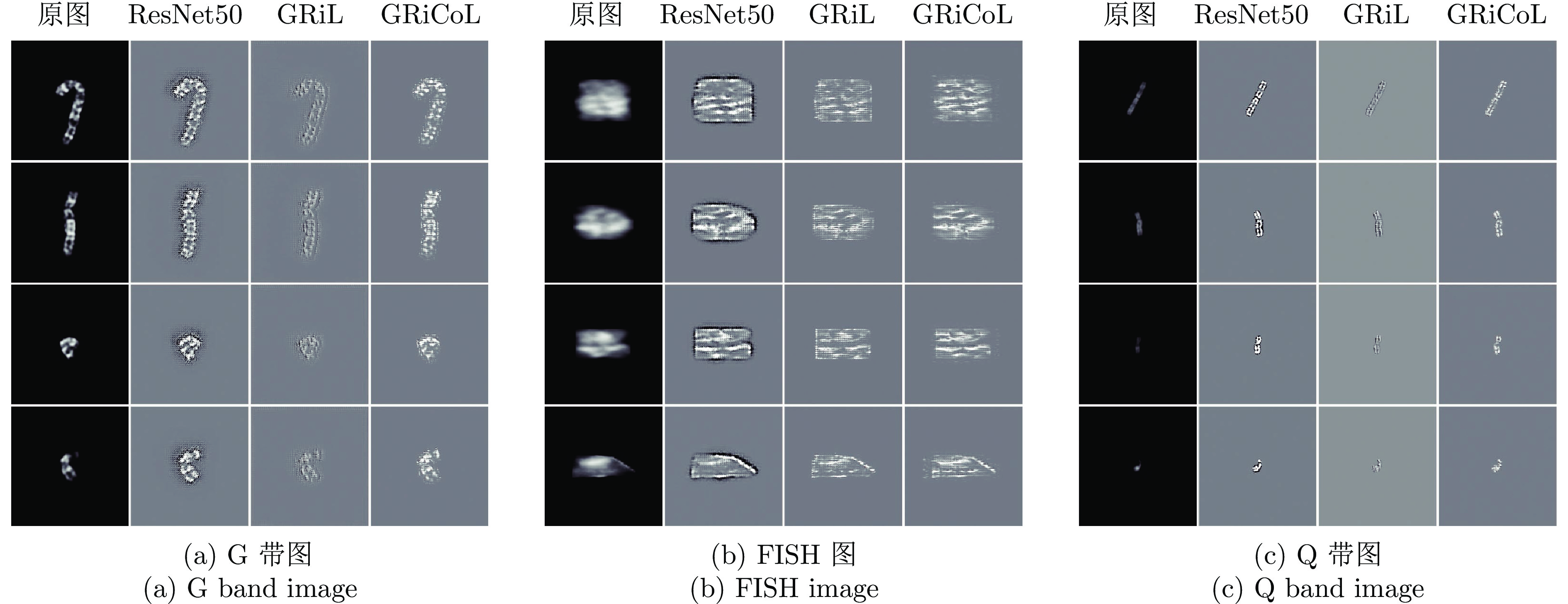
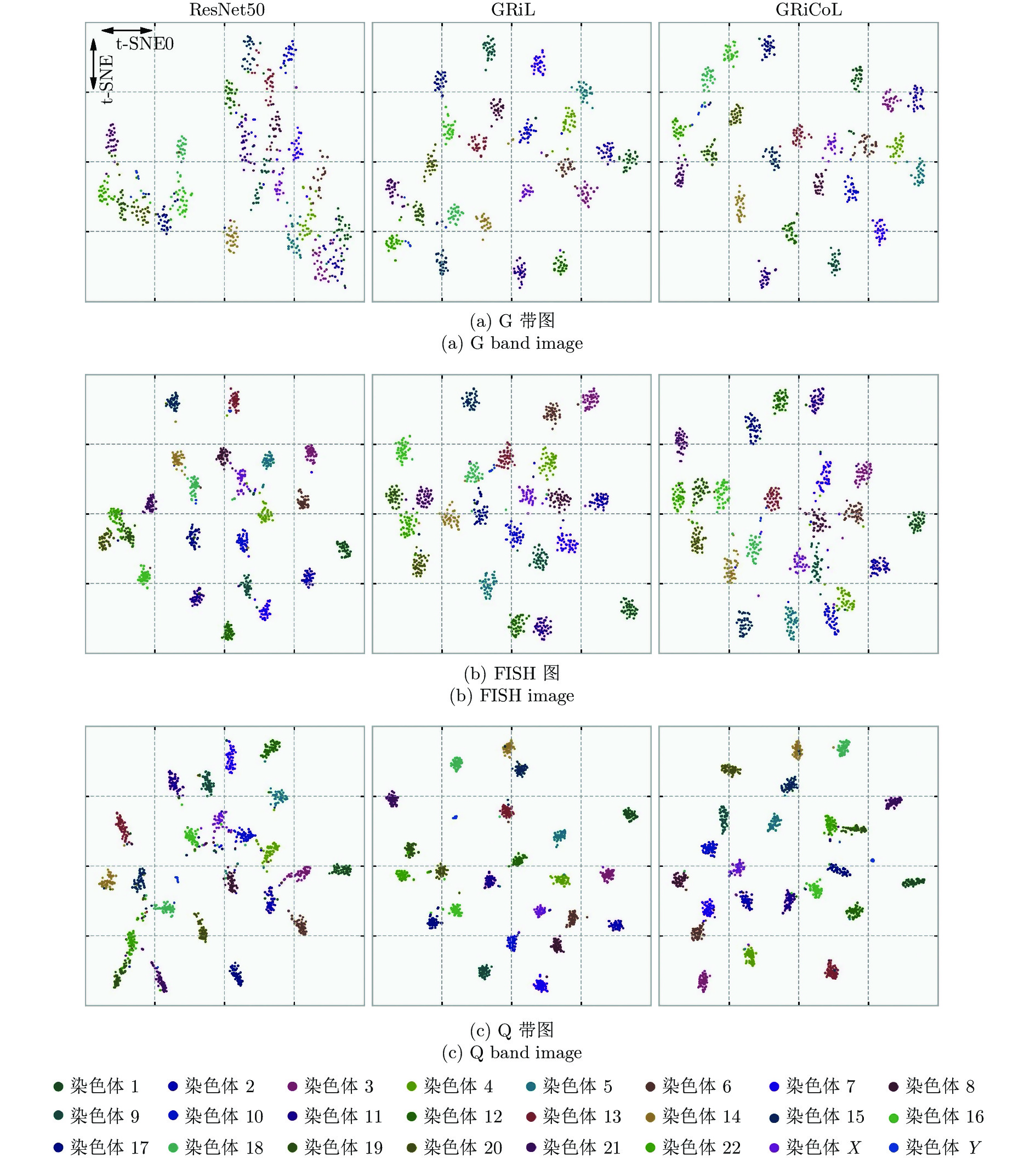
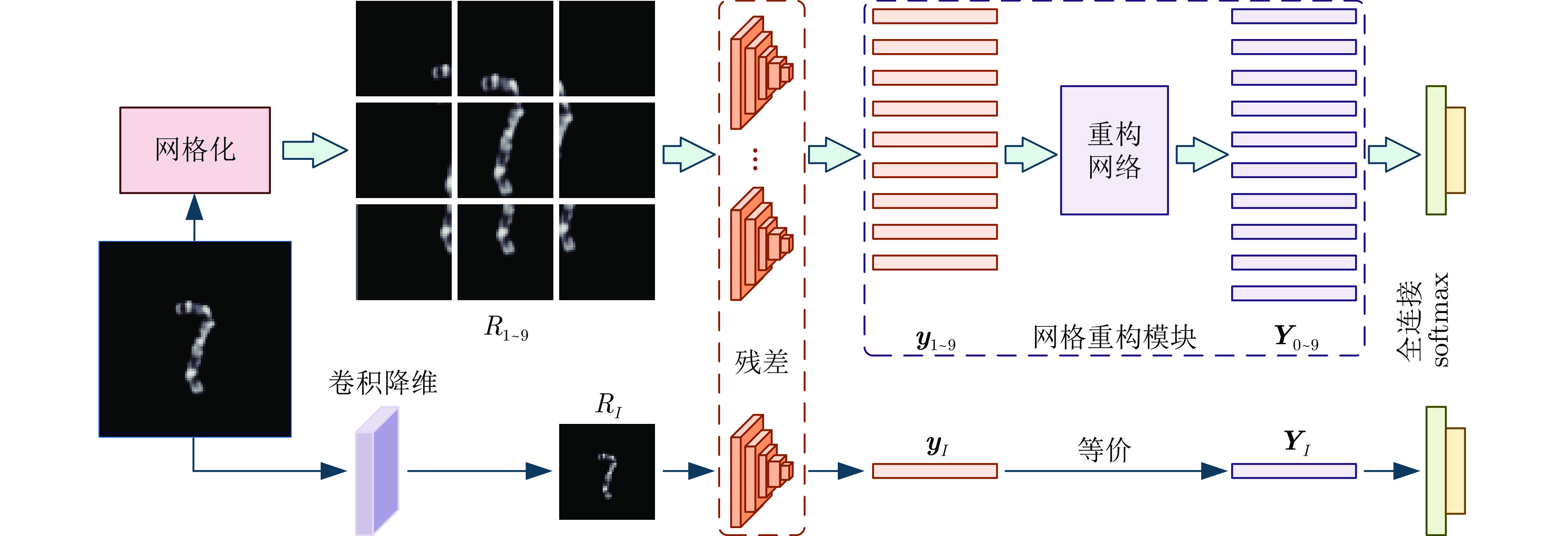
摘要: 染色体的分类是核型分析的重要任务之一. 因其柔软易弯曲, 且类间差异小、类内差异大等特点, 其精准分类仍然是一个具有挑战性的难题. 对此, 提出一种基于网格重构学习(Grid reconstruction learning, GRiCoL)的染色体分类模型. 该模型首先将染色体图像网格化, 提取局部分类特征; 然后通过重构网络对全局特征进行二次提取; 最后完成分类. 相比于现有几种先进方法, GRiCoL同时兼顾局部和全局特征提取更有效的分类特征, 有效改善染色体弯曲导致的分类性能下降, 参数规模合理. 通过基于G带、荧光原位杂交 (Fluorescence in situ hybridization, FISH)、Q带染色体公开数据集的实验表明: GRiCoL能够更好地弱化染色体弯曲带来的影响, 在不同数据集上的分类准确度均优于现有分类方法.Abstract: Chromosome classification is one of the key tasks of karyotype analysis. However, due to chromosomes are flexible hence exhibit less difference between different types while significant difference within same type, accurate classification of chromosome remains a challenging issue. In this paper, a chromosome classification model based on grid reconstruction learning (GRiCoL) is proposed. To weaken the impact of the bendy state, chromosome images are first grid-enabled for feature extraction separately. Subsequentially, global features are extracted for the second time by reconstruction network, which is followed by classification. Compared with the state-of-the-art methods, the proposed GRiCoL can get more efficient discriminable features based on both local and global features, therefore can overcome the adverse effects of bandy form of chromosome with reasonable parameter scale. Experiments on public G band, fluorescence in situ hybridization (FISH) as well as Q band chromosome datasets show that GRiCoL can extract discriminative features that weaken the bending of chromosomes more efficiently, meanwhile, higher performance was obtained as compared to current classification algorithms.
-
Key words:
- Karyotype analysis /
- chromosome classification /
- feature reconstruction /
- gridding
-
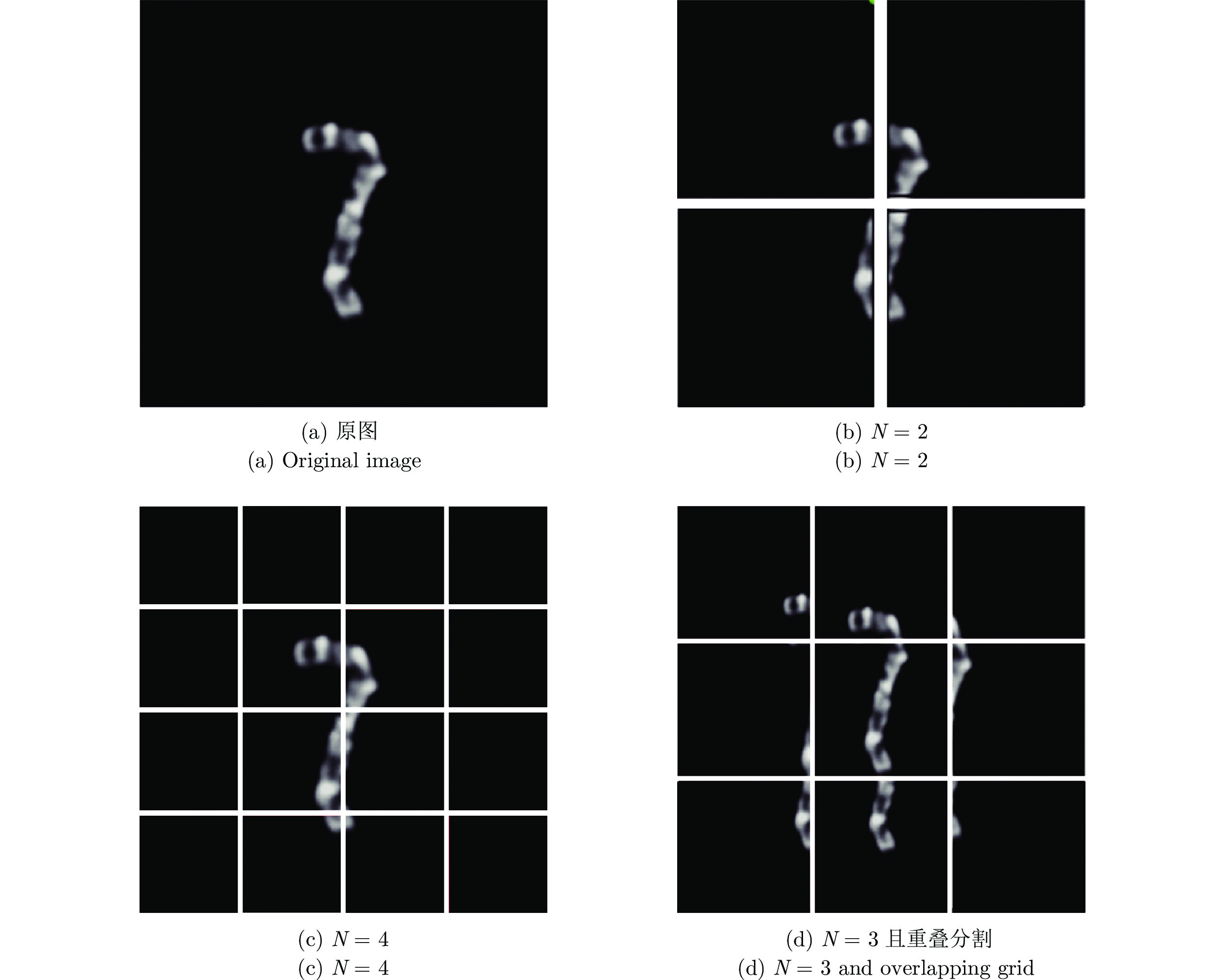
表 1 交叠网格设计的分类性能对比
Table 1 Classification performance comparison between grid with and without overlapping
模型 G带 FISH Q带 无交叠 GRiCoL 98.1% 96.2% 95.3% GRiCoL 99.5% 97.2% 97.3% p值 2.66e−22 0.52 1.71e−8 表 2 不同N数量下分类性能的对比
Table 2 Classification performance comparison between grids with different N
N G带(%) FISH (%) Q带(%) Gflops 参数量(M) 2 98.5 96.1 95.8 11.5 22.1 3 99.5 97.2 97.3 26.0 27.5 4 99.2 97.8 97.6 46.3 35.0 -
[1] Madian N, Jayanthi K B. Analysis of human chromosome classification using centromere position. Measurement, 2014, 47: 287−295 doi: 10.1016/j.measurement.2013.08.033 [2] Abid F, Hamami L. A survey of neural network based automated systems for human chromosome classification. Artificial Intelligence Review, 2018, 49(1): 41−56 doi: 10.1007/s10462-016-9515-5 [3] Poletti E, Grisan E, Ruggeri A. A modular framework for the automatic classification of chromosomes in Q-band images. Computer Methods and Programs in Biomedicine, 2012, 105(2): 120−130 doi: 10.1016/j.cmpb.2011.07.013 [4] Khan S, DSouza A, Sanches J, Ventura R. Geometric correction of deformed chromosomes for automatic Karyotyping. In: Proceedings of the Annual International Conference of the IEEE Engineering in Medicine and Biology Society. San Diego, USA: IEEE, 2012. 4438−4441 [5] Sharif Razavian A, Azizpour H, Sullivan J, Carlsson S. CNN features off-the-shelf: An astounding baseline for recognition. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition Workshops. Columbus, USA: IEEE, 2014. 512−519 [6] Lerner B, Guterman H, Dinstein I, Romem Y. Medial axis transform-based features and a neural network for human chromosome classification. Pattern Recognition, 1995, 28(11): 1673−1683 doi: 10.1016/0031-3203(95)00042-X [7] Ming D L, Tian J W. Automatic pattern extraction and classification for chromosome images. Journal of Infrared, Millimeter, and Terahertz Waves, 2010, 31(7): 866−877 doi: 10.1007/s10762-010-9640-1 [8] Markou C M C, Delopoulos A. Automatic chromosome classification using support vector machines. In: Proceedings of the IEEE Information Technology, Networking, Electronic and Automation Control Conference. 2012. 1−24 [9] Gao D Y, Madden M, Chambers D, Lyons G. Bayesian ANN classifier for ECG arrhythmia diagnostic system: A comparison study. In: Proceedings of the IEEE International Joint Conference on Neural Networks. Montreal, Canada: IEEE, 2005. 2383−2388 [10] 田娟秀, 刘国才, 谷珊珊, 鞠忠建, 刘劲光, 顾冬冬. 医学图像分析深度学习方法研究与挑战. 自动化学报, 2018, 44(3): 401−424Tian Juan-Xiu, Liu Guo-Cai, Gu Shan-Shan, Ju Zhong-Jian, Liu Jin-Guang, Gu Dong-Dong. Deep learning in medical image analysis and its challenges. Acta Automatica Sinica, 2018, 44(3): 401−424 [11] 宋燕, 王勇. 多阶段注意力胶囊网络的图像分类. 自动化学报, 2024, 50(9): 1804−1817Song Yan, Wang Yong. Multi-stage attention-based capsule networks for image classification. Acta Automatica Sinica, 2024, 50(9): 1804−1817 [12] 冯建周, 马祥聪. 基于迁移学习的细粒度实体分类方法的研究. 自动化学报, 2020, 46(8): 1759−1766Feng Jian-Zhou, Ma Xiang-Cong. Fine-grained entity type classification based on transfer learning. Acta Automatica Sinica, 2020, 46(8): 1759−1766 [13] Sharma M, Saha O, Sriraman A, Hebbalaguppe R, Vig L, Karande S. Crowdsourcing for chromosome segmentation and deep classification. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition Workshops (CVPRW). Honolulu, USA: IEEE, 2017. 786−793 [14] Swati, Gupta G, Yadav M, Sharma M, Vig L. Siamese networks for chromosome classification. In: Proceedings of the IEEE International Conference on Computer Vision Workshops (ICCVW). Venice, Italy: IEEE, 2017. 72−81 [15] Javan-Roshtkhari M, Setarehdan S K. A new approach to automatic classification of the curved chromosomes. In: Proceedings of the 5th International Symposium on Image and Signal Processing and Analysis. Istanbul, Turkey: IEEE, 2007. 19−24 [16] Zhang J P, Hu W J, Li S Y, Wen Y F, Bao Y, Huang H F, et al. Chromosome classification and straightening based on an interleaved and multi-task network. IEEE Journal of Biomedical and Health Informatics, 2021, 25(8): 3240−3251 doi: 10.1109/JBHI.2021.3062234 [17] Qin Y L, Wen J, Zheng H, Huang X L, Yang J, Song N, et al. Varifocal-net: A chromosome classification approach using deep convolutional networks. IEEE Transactions on Medical Imaging, 2019, 38(11): 2569−2581 doi: 10.1109/TMI.2019.2905841 [18] He K M, Zhang X Y, Ren S Q, Sun J. Deep residual learning for image recognition. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR). Las Vegas, USA: IEEE, 2016. 770−778 [19] Wu Y R, Yue Y S, Tan X, Wang W, Lu T. End-to-end chromosome karyotyping with data augmentation using GAN. In: Proceedings of the 25th IEEE International Conference on Image Processing (ICIP). Athens, Greece: IEEE, 2018. 2456−2460 [20] Yang Z, Luo T G, Wang D, Hu Z Q, Gao J, Wang L W. Learning to navigate for fine-grained classification. In: Proceedings of the 15th European Conference on Computer Vision (ECCV). Munich, Germany: Springer, 2018. 438−454 [21] 罗建豪, 吴建鑫. 基于深度卷积特征的细粒度图像分类研究综述. 自动化学报, 2017, 43(8): 1306−1318Luo Jian-Hao, Wu Jian-Xin. A survey on fine-grained image categorization using deep convolutional features. Acta Automatica Sinica, 2017, 43(8): 1306−1318 [22] Cui Y, Zhou F, Wang J, Liu X, Lin Y Q, Belongie S. Kernel pooling for convolutional neural networks. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR). Honolulu, USA: IEEE, 2017. 3049−3058 [23] Lin T Y, RoyChowdhury A, Maji S. Bilinear CNN models for fine-grained visual recognition. In: Proceedings of the IEEE International Conference on Computer Vision (ICCV). Santiago, Chile: IEEE, 2015. 1449−1457 [24] Chen Y, Bai Y L, Zhang W, Mei T. Destruction and construction learning for fine-grained image recognition. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR). Long Beach, USA: IEEE, 2019. 5152−5161 [25] Fu J L, Zheng H L, Mei T. Look closer to see better: Recurrent attention convolutional neural network for fine-grained image recognition. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR). Honolulu, USA: IEEE, 2017. 4476−4484 [26] Wei X S, Xie C W, Wu J X, Shen C H. Mask-CNN: Localizing parts and selecting descriptors for fine-grained bird species categorization. Pattern Recognition, 2018, 76: 704−714 doi: 10.1016/j.patcog.2017.10.002 [27] Fukui H, Hirakawa T, Yamashita T, Fujiyoshi H. Attention branch network: Learning of attention mechanism for visual explanation. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR). Long Beach, USA: IEEE, 2019. 10697−10706 [28] Lin C C, Zhao G S, Yang Z R, Yin A H, Wang X M, Guo L, et al. CIR-Net: Automatic classification of human chromosome based on inception-ResNet architecture. IEEE/ACM Transactions on Computational Biology and Bioinformatics, 2022, 19(3): 1285−1293 doi: 10.1109/TCBB.2020.3003445 [29] Hu X, Yi W L, Jiang L, Wu S J, Zhang Y, Du J Q, et al. Classification of metaphase chromosomes using deep convolutional neural network. Journal of Computational Biology, 2019, 26(5): 473−484 doi: 10.1089/cmb.2018.0212 [30] Wang X W, Zheng B, Li S B, Mulvihill J J, Wood M C, Liu H. Automated classification of metaphase chromosomes: Optimization of an adaptive computerized scheme. Journal of Biomedical Informatics, 2009, 42(1): 22−31 doi: 10.1016/j.jbi.2008.05.004 [31] 刘嘉敏, 苏远歧, 魏平, 刘跃虎. 基于长短记忆与信息注意的视频−脑电交互协同情感识别. 自动化学报, 2020, 46(10): 2137−2147Liu Jia-Min, Su Yuan-Qi, Wei Ping, Liu Yue-Hu. Video-EEG based collaborative emotion recognition using LSTM and information-attention. Acta Automatica Sinica, 2020, 46(10): 2137−2147 [32] Springenberg J T, Dosovitskiy A, Brox T, Riedmiller M A. Striving for simplicity: The all convolutional net. In: Proceedings of the 3rd International Conference on Learning Representations. San Diego, USA: 2015. [33] Van der Maaten L, Hinton G. Visualizing data using t-SNE. Journal of Machine Learning Research, 2008, 9: 2579−2605 [34] Zhang Y Z, Liu S W, Qi L, Coleman S, Kerr D, Shi W D. Multi-level and multi-scale horizontal pooling network for person re-identification. Multimedia Tools and Applications, 2020, 79(39): 28603−28619 [35] 王雅湄, 王振友. 基于多区域特征的深度卷积神经网络模型. 应用数学进展, 2019, 8(11): 1753−1765 doi: 10.12677/AAM.2019.811205Wang Ya-Mei, Wang Zhen-You. Deep convolutional neural network model based on multi-region feature. Advances in Applied Mathematics, 2019, 8(11): 1753−1765 doi: 10.12677/AAM.2019.811205 -




 下载:
下载: